Scanning¶
A regular tomographic scan at 2-BM consists of a series of projections collected in fly scan mode and a set of dark/flat images collected before/after/both the projections. These tasks are accomplished by tomoScan. 2-BM can also collect tomographic data in streaming mode. In this case projections and dark/flat images are broadcasted as EPICS PVs and the actual data saving only occurs on-demand. The data collected in streaming mode is accomplished by tomoScanStream. During the streaming data collection, a real-time 3-orthogonal slice reconstruction is also generated using tomoStream
Since at 2-BM we have two startion (A and B), tomoScan and tomoStream are installed at both station. To access the main control screens
[user2bmb@arcturus]$ start_epics
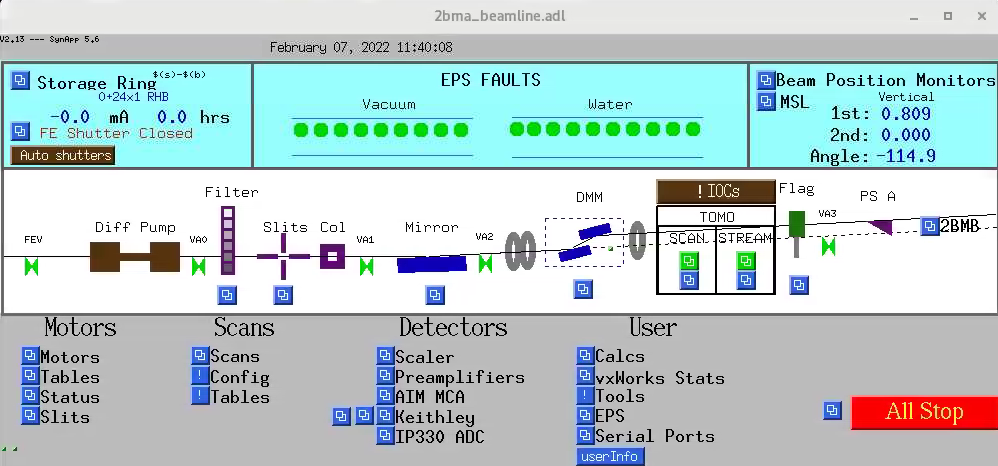
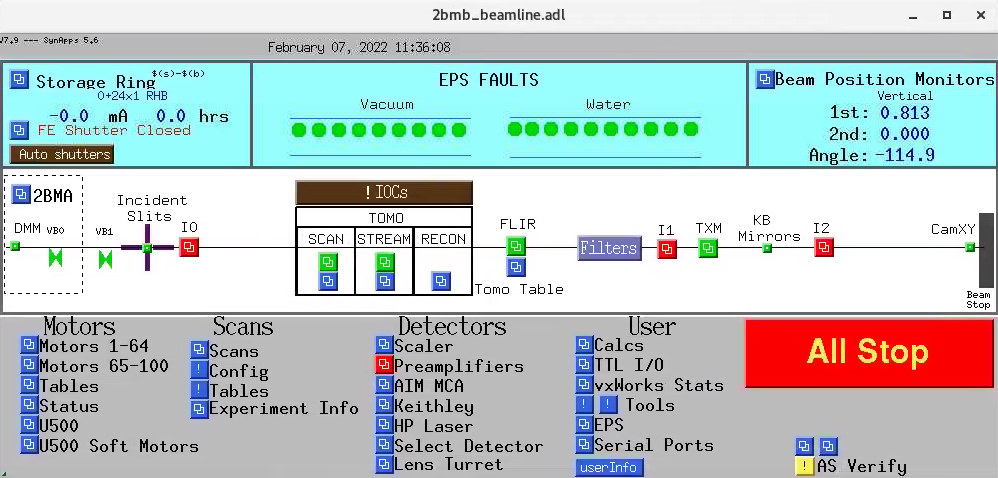
To start the main tomography control screens for 2-BM-A or 2-BM-B for scanning or streaming data collection select in the main beamline control screens the corresponding screen for user, admin, tomoscan, tomoscan 2-BM:
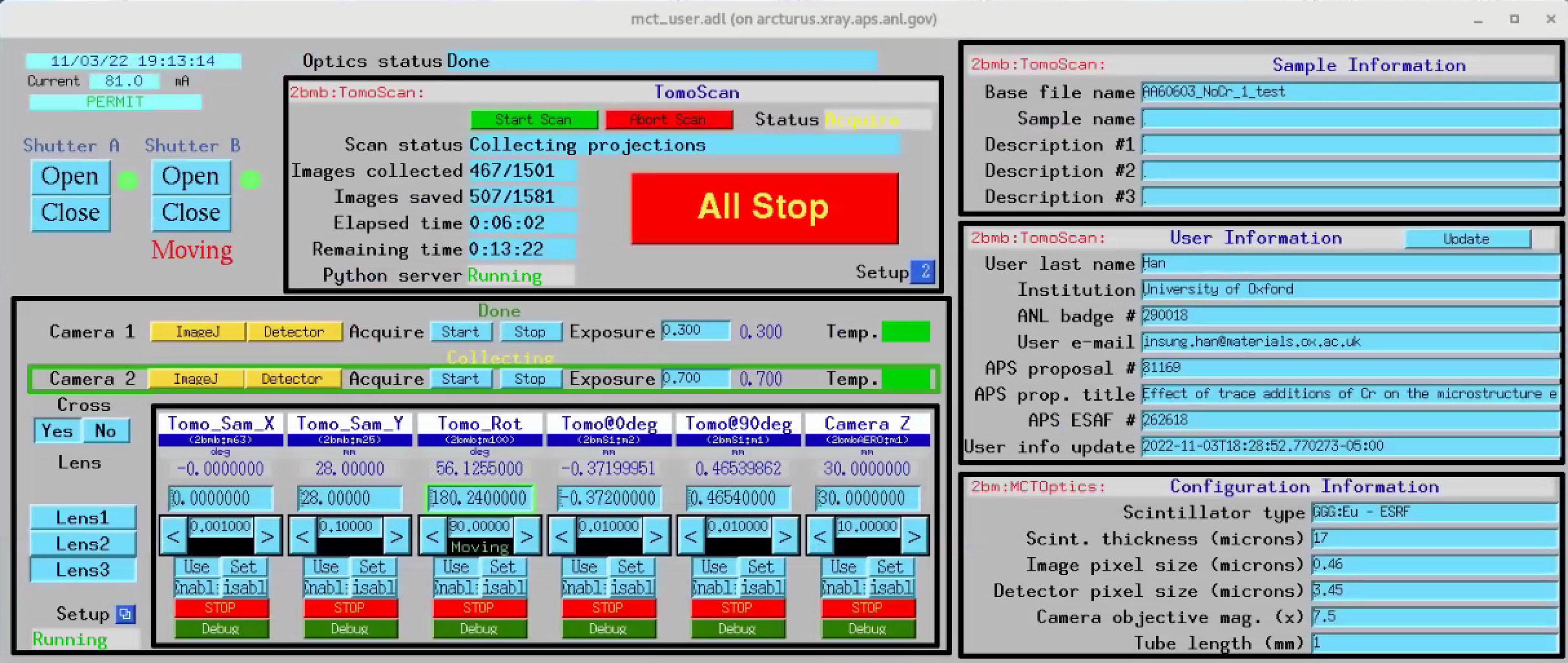
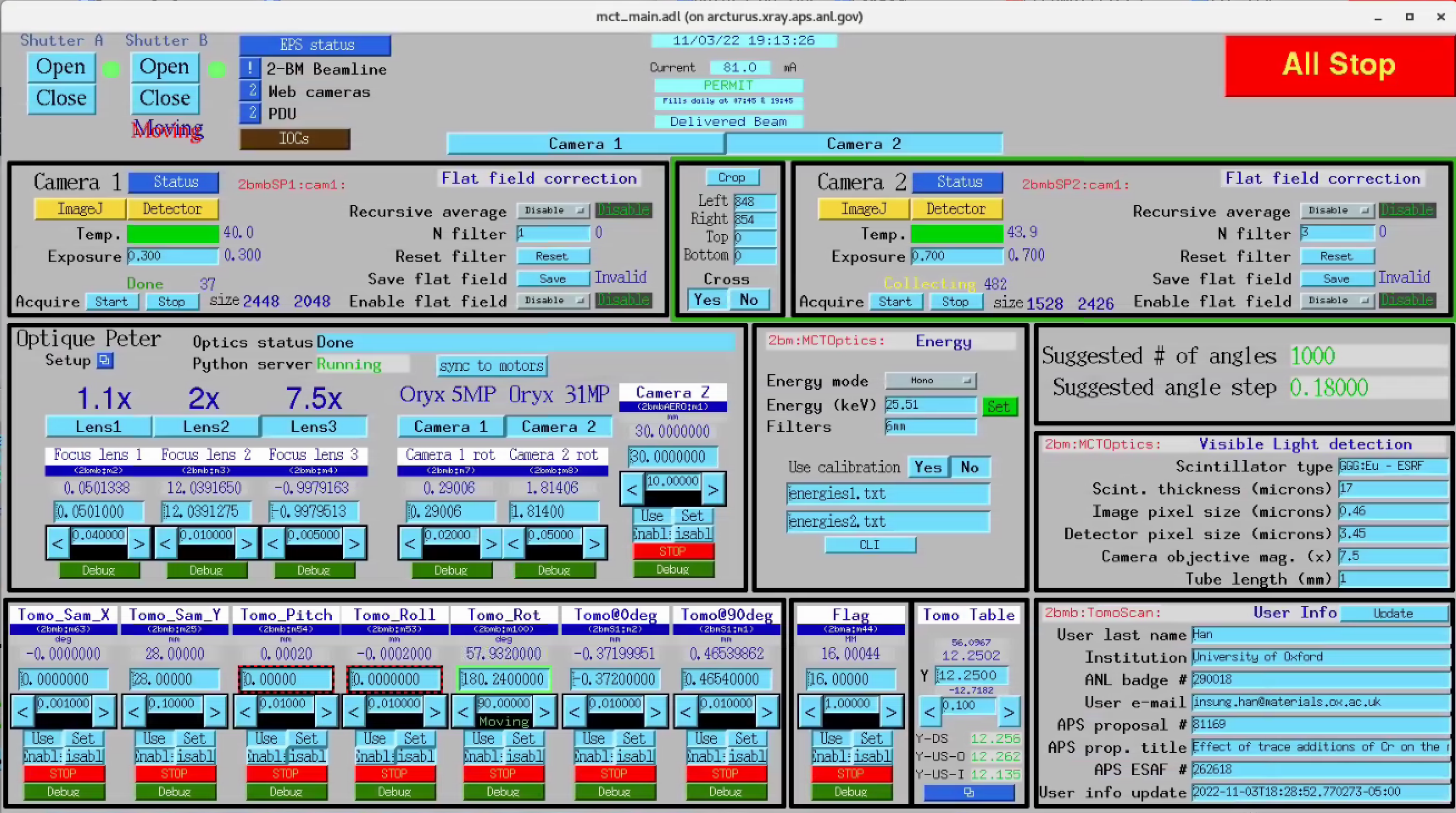
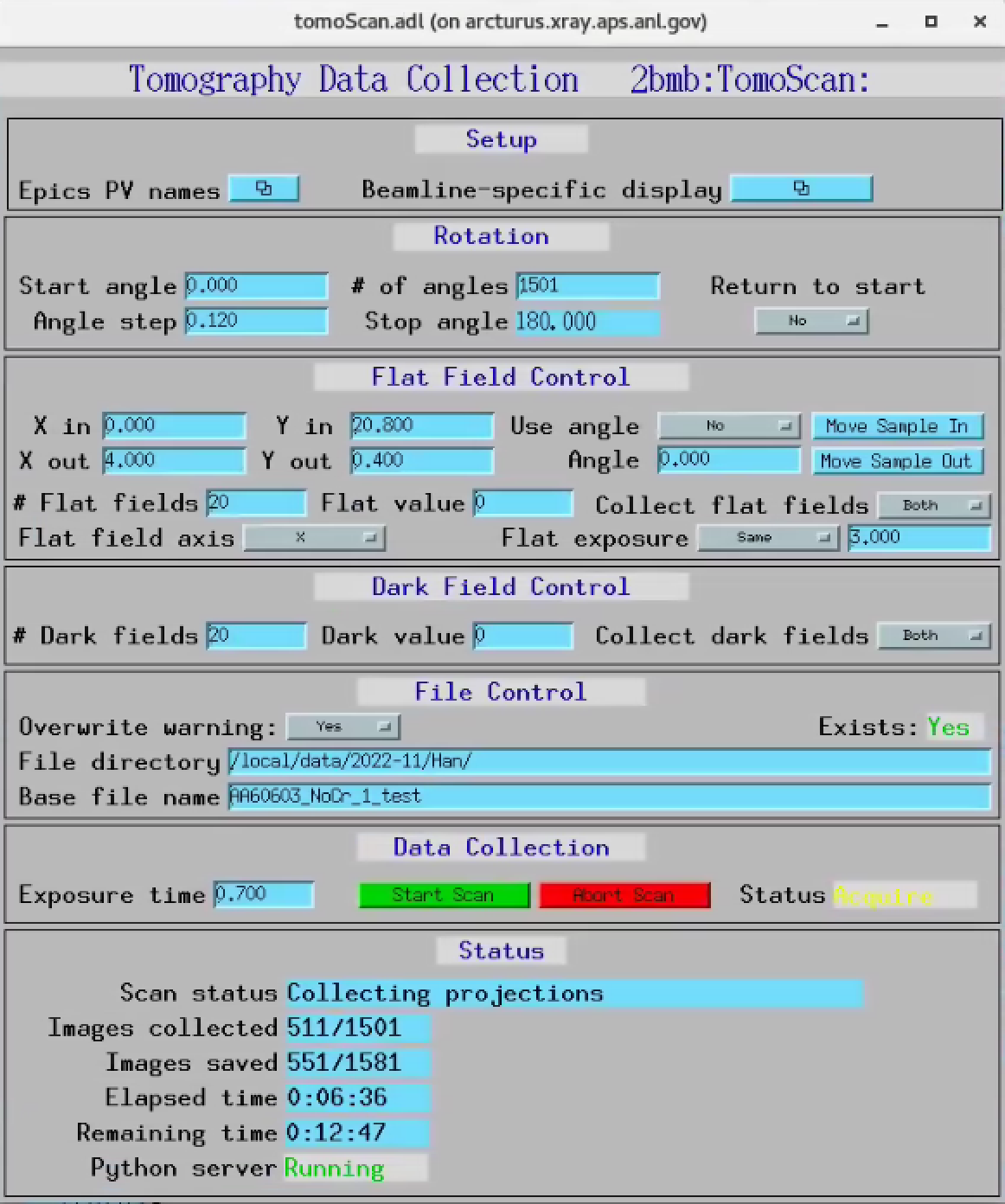
TomoScan¶
There are 2 installation of tomoScan at 2-BM to support tomography in the A and B stations.
Startup¶
For experiments in 2-BM-A:
[user2bmb@arcturus]$ ~/tomoscan_start_A.sh
for experiments in 2-BM-B:
[user2bmb@arcturus]$ ~/tomoscan_start_B.sh
a_tomoscan_start.sh and b_tomoscan_start.sh for scripts content.
The tomoscan startup steps executed by a_tomoscan_start.sh for experiments in 2-BM-A are described in detail below.
Data collection¶
Support for tomography data collection is provided by tomoScan_2bm a tomoScan derived classes to implement the data collection at 2-BM. To run tomoScan in 2-BM-A:
Start area detector - EPICS IOC
[user2bmb@pg10ge]$ 2bmSP2 start
medm screen
[user2bmb@pg10ge]$ 2bmSP2 medm
Start tomoScan
EPICS IOC
[user2bmb@pg10ge]$ cd /local/user2bmb/epics/synApps/support/tomoscan/iocBoot/iocTomoScan_2BMA/
[user2bmb@pg10ge]$ ./start_IOC
tomoscan_2bm python server
[user2bmb@pg10ge]$ bash
[user2bmb@pg10ge]$ cd /local/user2bmb/epics/synApps/support/tomoscan/iocBoot/iocTomoScan_2BMA/
[user2bmb@pg10ge]$ python -i start_tomoscan.py
medm screen
[user2bmb@pg10ge]$ cd /local/user2bmb/epics/synApps/support/tomoscan/iocBoot/iocTomoScan_2BMA/
[user2bmb@pg10ge]$ ./start_medm
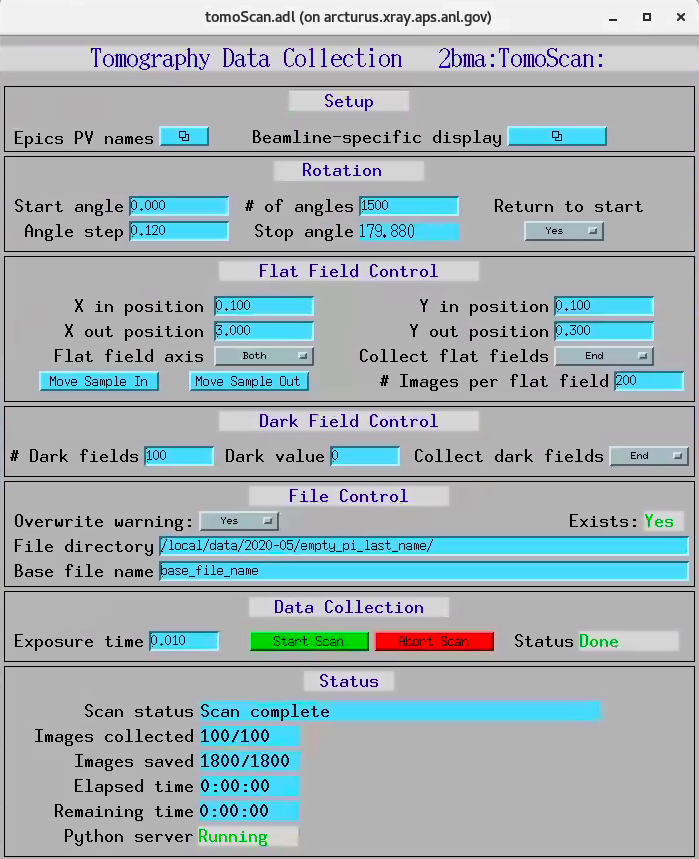
The tomoScan allows to configure and collect a single tomographic dataset.
Command-line-interface
[user2bmb@pg10ge]$ tomoscan -h
usage: tomoscan [-h] [--config FILE] [--version] ...
optional arguments:
-h, --help show this help message and exit
--config FILE File name of configuration file
--version show program's version number and exit
Commands:
init Create configuration file
status Show tomoscan status
single Run a single tomographic scan
vertical Run a vertical tomographic scan
horizontal Run a horizontal tomographic scan
mosaic Run a mosaic tomographic scan
each command help is accessible with -h:
Usage: tomoscan vertical [-h] [--scan-type SCAN_TYPE]
[--tomoscan-db-home FILE]
[--tomoscan-prefix TOMOSCAN_PREFIX]
[--in-situ-pv IN_SITU_PV]
[--in-situ-pv-rbv IN_SITU_PV_RBV]
[--in-situ-start IN_SITU_START]
[--in-situ-step-size IN_SITU_STEP_SIZE]
[--sleep-steps SLEEP_STEPS] [--sleep-time SLEEP_TIME]
[--vertical-start VERTICAL_START]
[--vertical-step-size VERTICAL_STEP_SIZE]
[--vertical-steps VERTICAL_STEPS] [--config FILE]
[--in-situ] [--logs-home FILE] [--sleep] [--testing]
[--verbose]
optional arguments:
-h, --help show this help message and exit
--scan-type SCAN_TYPE
For internal use to log the tomoscan status (default: )
--tomoscan-db-home FILE
Log file directory
(default: /home/user2bmb/epics/synApps/support/tomoscan/db/)
--tomoscan-prefix TOMOSCAN_PREFIX
The tomoscan prefix, i.e.'13BMDPG1:TS:' or
'2bma:TomoScan:' (default: 2bma:TomoScan:)
--in-situ-pv IN_SITU_PV
Name of the in-situ EPICS process variable to set
(default: )
--in-situ-pv-rbv IN_SITU_PV_RBV
Name of the in-situ EPICS process variable to read back (default: )
--in-situ-start IN_SITU_START
In-situ start (default: 0)
--in-situ-step-size IN_SITU_STEP_SIZE
In-situ step size (default: 1)
--sleep-steps SLEEP_STEPS
Number of sleep/in-situ steps (default: 1)
--sleep-time SLEEP_TIME
Wait time (s) between each data collection scan (default: 0)
--vertical-start VERTICAL_START
Vertical start position (mm) (default: 0)
--vertical-step-size VERTICAL_STEP_SIZE
Vertical step size (mm) (default: 1)
--vertical-steps VERTICAL_STEPS
Number of vertical steps (default: 1)
--config FILE File name of configuration file
(default: /home/user2bmb/tomoscan.conf)
--in-situ Enable in-situ PV scan during sleep time (default: False)
--logs-home FILE Log file directory (default: /home/user2bmb/logs)
--sleep Enable sleep time between tomography scans (default: False)
--testing Enable test mode, tomography scan will not run (default: False)
--verbose Verbose output (default: False)
to run a single scan with the parameters set in the tomoScan IOC and the tomoscan-cli:
[user2bmb@pg10ge]$ tomoscan single
tomoscan supports also vertical, horizontal and mosaic tomographic scans with:
[user2bmb@pg10ge]$ tomoscan vertical
[user2bmb@pg10ge]$ tomoscan horizontal
[user2bmb@pg10ge]$ tomoscan mosaic
to run a vertical scan:
$ [user2bmb@pg10ge]$ tomoscan vertical --vertical-start 0 --vertical-step-size 0.1 --vertical-steps 2
2020-05-29 16:54:03,354 - vertical scan start
2020-05-29 16:54:03,356 - vertical positions (mm): [0. 0.1]
2020-05-29 16:54:03,358 - SampleInY stage start position: 0.000 mm
2020-05-29 16:54:03,362 - single scan start
2020-05-29 16:54:51,653 - single scan time: 0.805 minutes
2020-05-29 16:54:51,654 - SampleInY stage start position: 0.100 mm
2020-05-29 16:54:51,658 - single scan start
2020-05-29 16:55:47,607 - single scan time: 0.932 minutes
2020-05-29 16:55:47,607 - vertical scan time: 1.738 minutes
2020-05-29 16:55:47,608 - vertical scan end
tomoscan-cli always stores the last used set of paramters so to repeat the above vertical scan:
[user2bmb@pg10ge]$ tomoscan vertical
use -h for the list of supported parameters.
To repeat the vertical scan 5 times with 60 s wait time between each:
[user2bmb@pg10ge]$ tomoscan vertical --sleep --sleep-steps 10 --sleep-time 60
to repeat the same:
[user2bmb@pg10ge]$ tomoscan vertical --sleep
while:
[user2bmb@pg10ge]$ tomoscan vertical
repeats a single vertical scan with –vertical-start 0 –vertical-step-size 0.1 –vertical-steps 5.
To reset the tomoscan-cli status:
[user2bmb@pg10ge]$ tomoscan init
after deleting the tomoscan.conf file if already exists.
TomoStream¶
There are 2 major components supporting streaming at 2-BM:
Streaming data collection
Streaming data reconstruction
Startup¶
To start streaming data collection and streaming data reconstruction run tomostream_start.sh,
for experiments in 2-BM-A:
[tomo@handyn]$ ~/tomostream_start.sh
for experiments in 2-BM-B:
[user2bmb@arcturus]$ ~/tomostream_start.sh
a_tomostream_start.sh and b_tomostream_start.sh for scripts content.
The streaming data collection and streaming data reconstruction startup steps executed by b_tomostream_start.sh for experiments in 2-BM-B are described in detail below.
Streaming data collection¶
Support for streaming data collection is provided by tomoScanStream a tomoScan derived classes to implement the streaming data collection. To run tomoScanStream in 2-BM-B:
Start area detector
EPICS IOC
[user2bmb@lyra]$ 2bmbSP2 start
[user2bmb@lyra]$ 2bmbSP2 console
medm screen
[user2bmb@lyra]$ 2bmbSP2 medm
Note: the IOC prefix for [user2bmb@lyra]$ 2bmbSP2 start is 2bmbSP1:
Start tomoScanStream
EPICS IOC
[user2bmb@lyra]$ cd /local/user2bmb/epics/synApps/support/tomoscan/iocBoot/iocTomoScanStream_2BMB/
[user2bmb@lyra]$ ./start_IOC
tomoscan_stream python server
[user2bmb@lyra]$ bash
[user2bmb@lyra]$ cd /local/user2bmb/epics/synApps/support/tomoscan/iocBoot/iocTomoScanStream_2BMB/
[user2bmb@lyra]$ python -i start_tomoscan_stream.py
medm screen
[user2bmb@lyra]$ cd /local/user2bmb/epics/synApps/support/tomoscan/iocBoot/iocTomoScanStream_2BMB/
[user2bmb@lyra]$ ./start_medm
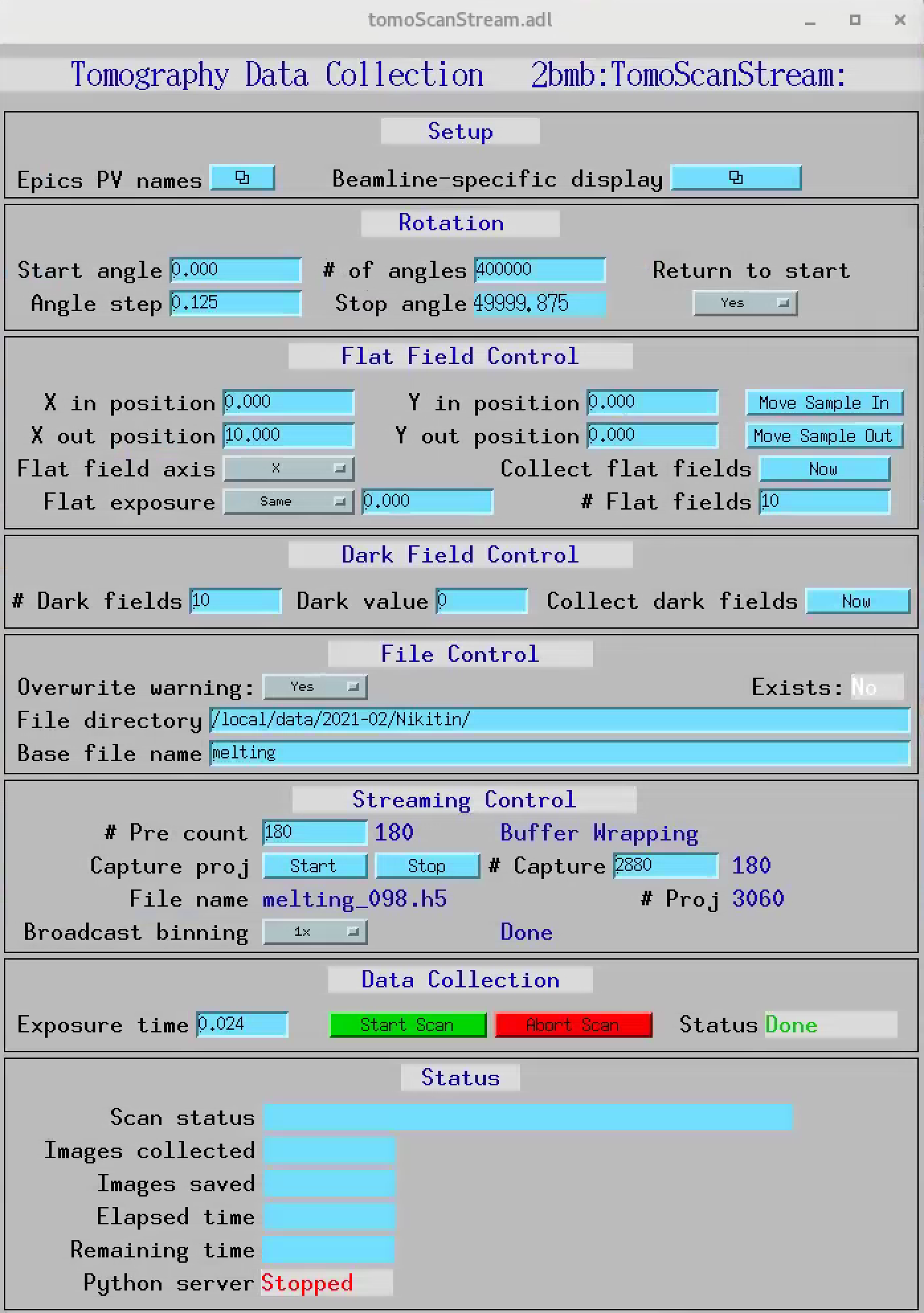
Streaming data collection features can be controlled from the Streaming Control section and includes:
On-demand data capturing with saving in a standard Data Exchange hdf5file
Set a number of projectons (“Pre count”) collected before a triggered data capturing event to be also saved in the same hdf5 file
binning data streaming
dark-flat field images can be re-taken on-demand at any time during data collection by selecting Now next to the Collect flat (dark) fields.
When collecting data in streaming mode, projections, dark and flat images are broadcasted using PVaccess and can be retrieved as EPICS PVs. Projections are streamed by the detector PVA1 plugin while dark and flat are streamed by tomoScanStream with a dark/flat Stream Prefix configurable under tomoScan/Epics PV names PVs screen:
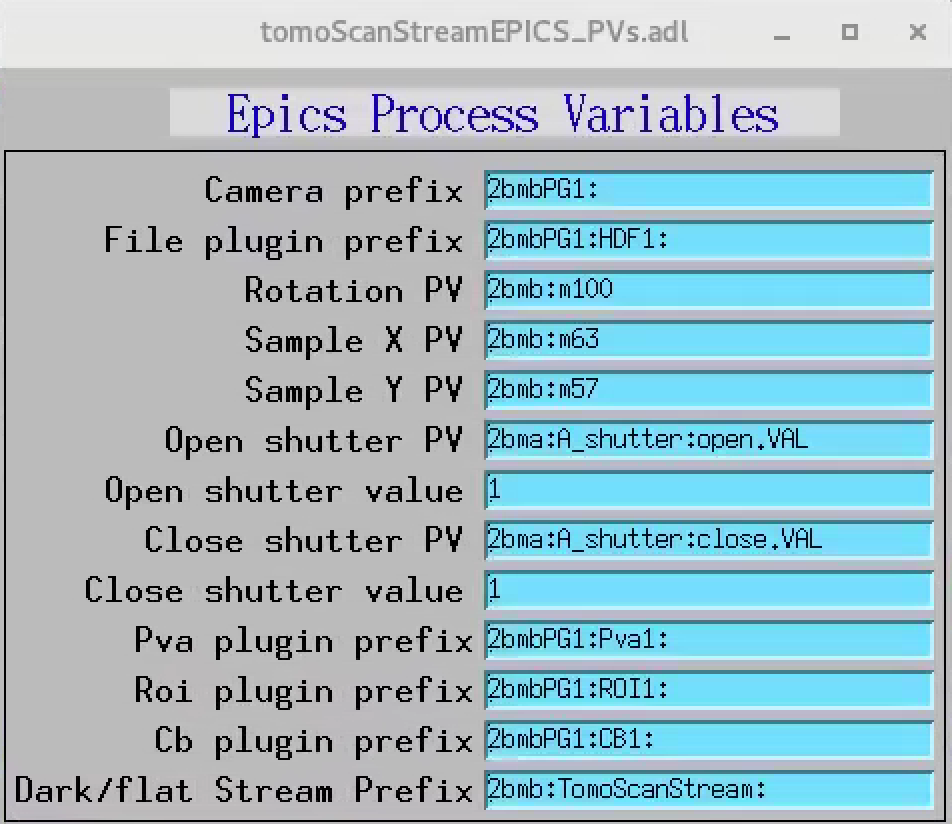
Using the dark/flat Stream Prefix above, the PVs for data and flat are:
2bmb:TomoScanStream:flat
2bmb:TomoScanStream:dark
These PVs together with the projection PV (in this case 2bmbSP1:Pva1:) will be passed as input to the tomography streaming reconstruction tool tomoStream.
Streaming data reconstruction
The projection, dark and flat image broadcast provided by tomoScanStream can be used to reconstruct in real-time 3 orthogonal slices. This task is accomplished by tomoStream.
Start tomoStream
EPICS IOC
[tomo@handyn]$ cd /local/tomo/epics/synApps/support/tomostream/iocBoot/iocTomoStream/
[tomo@handyn]$ ./start_IOC
tomostream python server
[tomo@handyn]$ bash
[tomo@handyn]$ cd /local/tomo/epics/synApps/support/tomostream/iocBoot/iocTomoStream/
[tomo@handyn]$ source activate streaming
[tomo@handyn]$ python -i start_tomostream.py
medm screen
[tomo@handyn]$ bash
[tomo@handyn]$ cd /local/tomo/epics/synApps/support/tomostream/iocBoot/iocTomoStream/
[tomo@handyn]$ ./start_medm
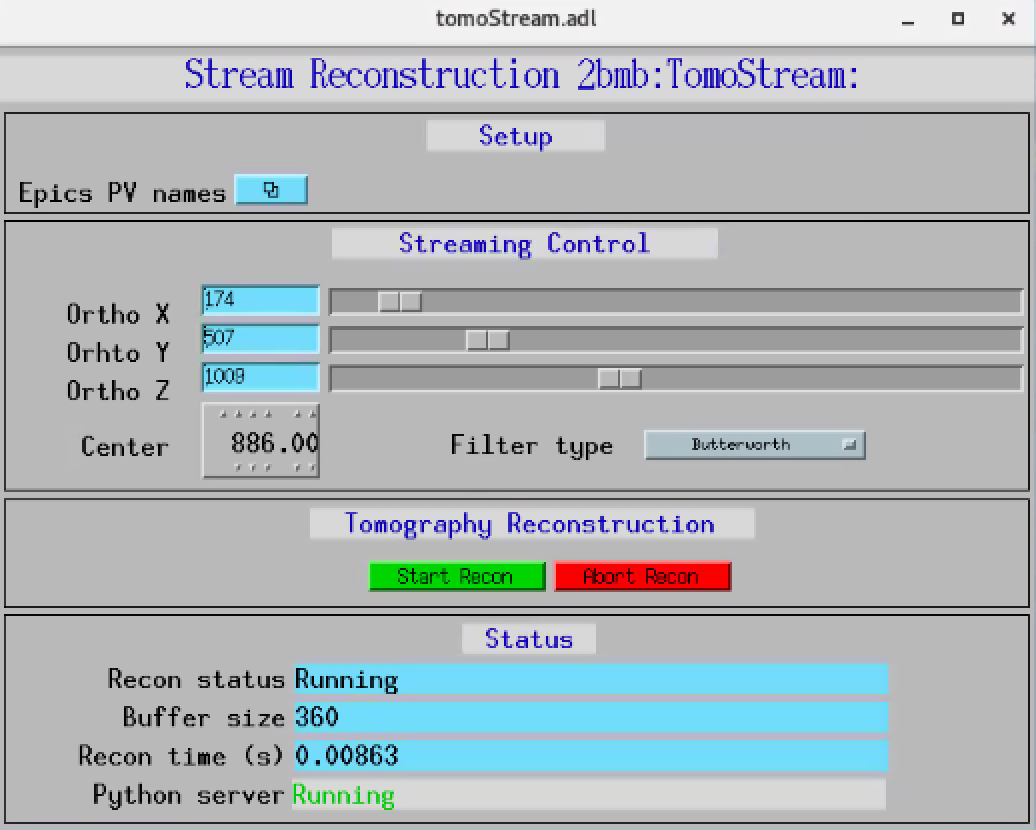
Streaming data reconstruction features are:
Streaming reconstruction of 3 (X-Y-Z) ortho-slices through the sample
On demand adjustment of the
X Y Z ortho-slice positions
reconstruction rotation center
reconstruction filter
and can be controlled from the main tomoStream control screen.
The output of tomostream is a live reconstruction:
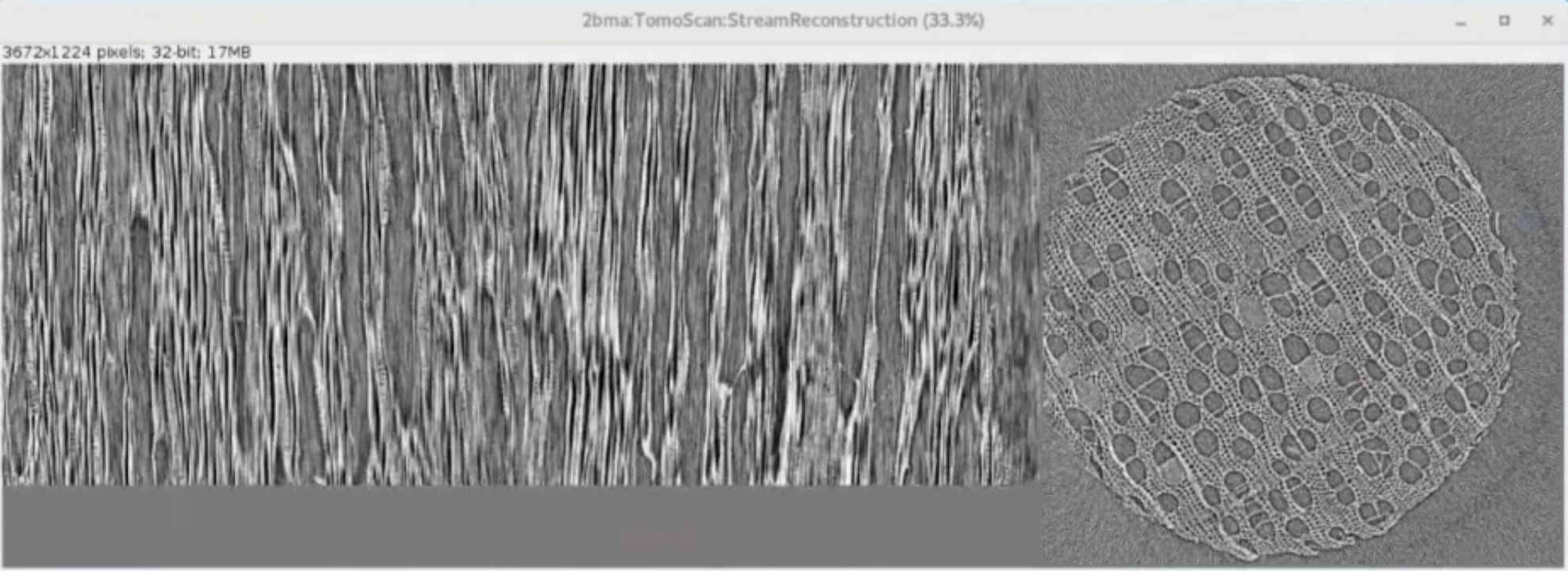
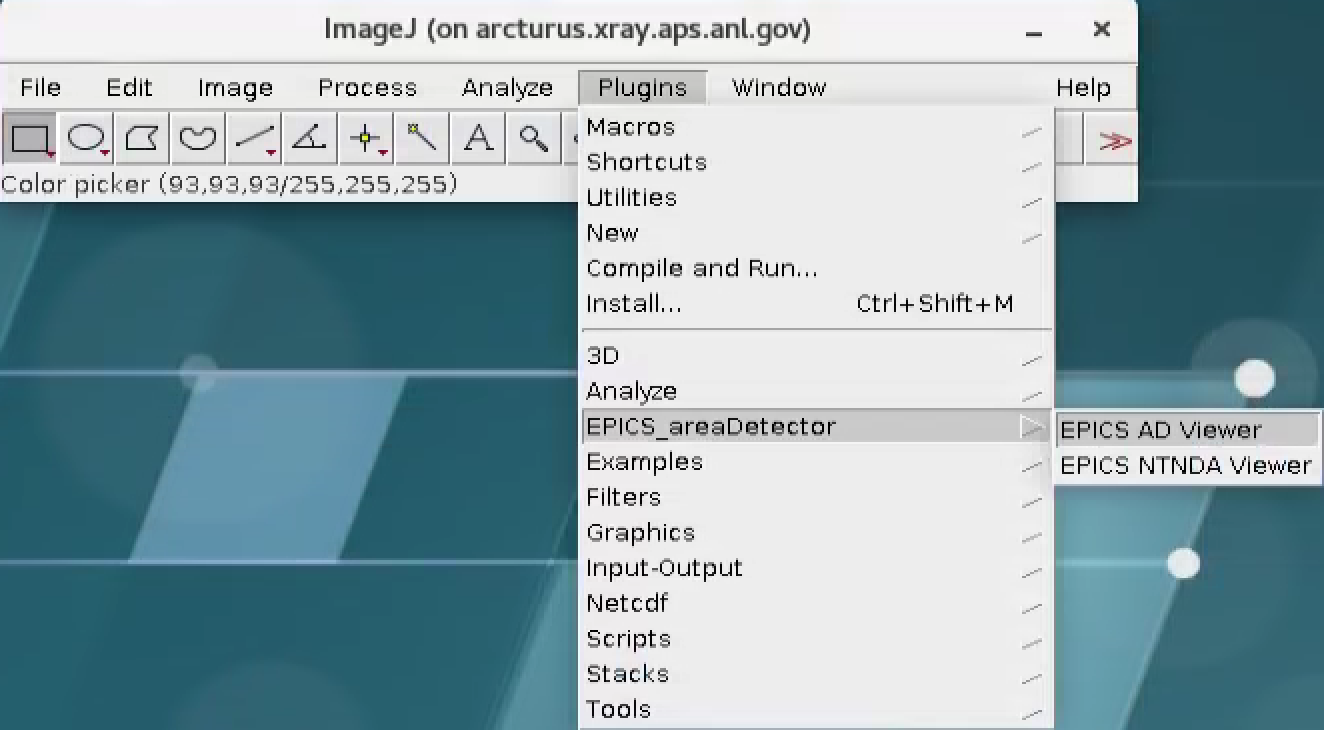
This is broadcasted as a PVA that can be diplayed by ImageJ using the EPICS_NTNDA_Viewer plug-in:
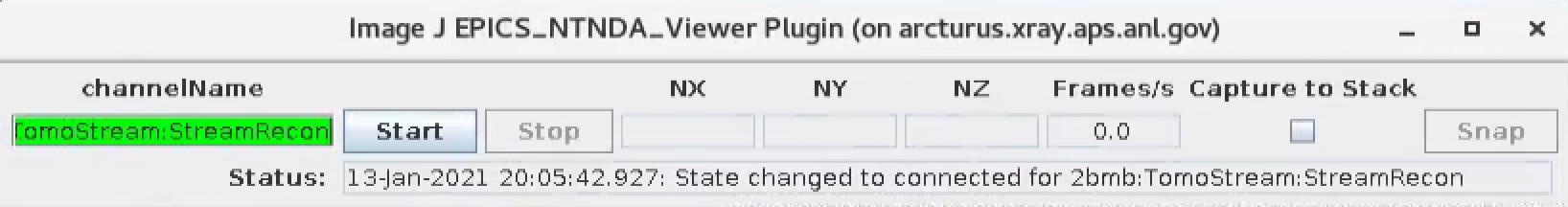
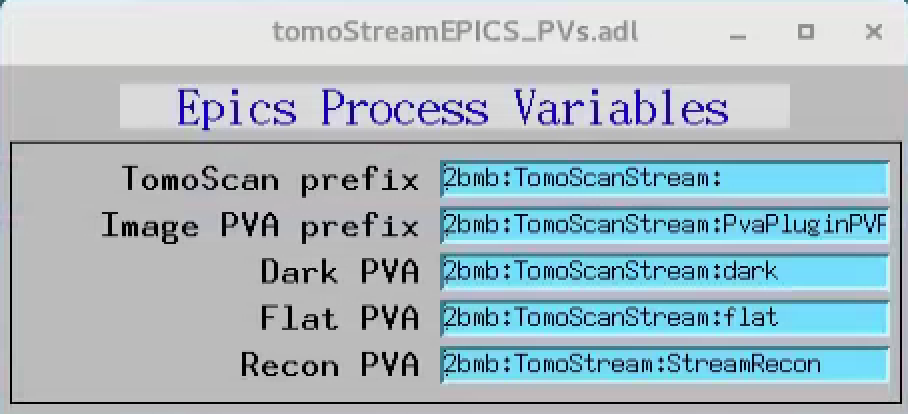
The PVA name broadcasting the recontruction can be set in the tomoStream/Epics PV names under Recon PVA screen:
While the sample is rotating is possible to optimize instrument (alignment, focus, sample to detector distance etc.) and beamline (energy etc.) conditions and monitor the effect live on the 3 orthogonal slices. It is also possible to automatically trigger data capturing based on events occurring in the sample and its environment as a result of segmentation or machine learning.